This project attempted to improve the quality of OCR applied to difficult entomology images[*] by cropping labels from the images to run through OCR separately. In order to identify labels on the image to crop, an initial, 'naive' pass of OCR was made over the whole image, generating both
- A) a set of rectangles on the image defined as word bounding boxes by the OCR engine, and
- B) a control OCR text file to be used for comparing the 'naive' model with the methodology.
Those word rectangles were then filtered, consolidated, and filtered again to identify the labels on the image, which were then extracted and run through the OCR engine separately. The resulting OCR output files were then concatenated into a single text file, which was compared against the 'naive' output described in A (above).
I'll call this method "ocrocrop". (For more detail on method, see the transcript of my preliminary presentation.)
The results were encouraging. (See CSV file listing results for each file, and the directory containing "naive" output, annotated JPGs, and cleaned output files for each test.)
Of 80 files tested, 20 experienced a decrease in score (see Alex Thomson's scoring service), but most (14/20) of those were on OCR output below 10% accuracy in the first place, and the remainder were at or below 20% accuracy. So it is reasonable to say that the ocrocrop method only degraded the quality of texts that were unusable in the first place.
40 of the 80 files tested showed more promising results, showing improvements from one to twenty percentage points -- in some cases only marginally improving unusable (below 10% accurate) outputs, but in many cases improving the scores more substantially (say from 25% to 35% in the case of EMEC609908_Stigmus_sp).
Most of the top quartile of results saw improvements on texts that were already scoring above 10% accuracy rates (16 of 20), so it appears that the effectiveness of the ocrocrop method is correlated to the quality of the naive input data -- garbage is degraded or only minimally improved, while OCR that is merely bad under the naive approach can be significantly improved.
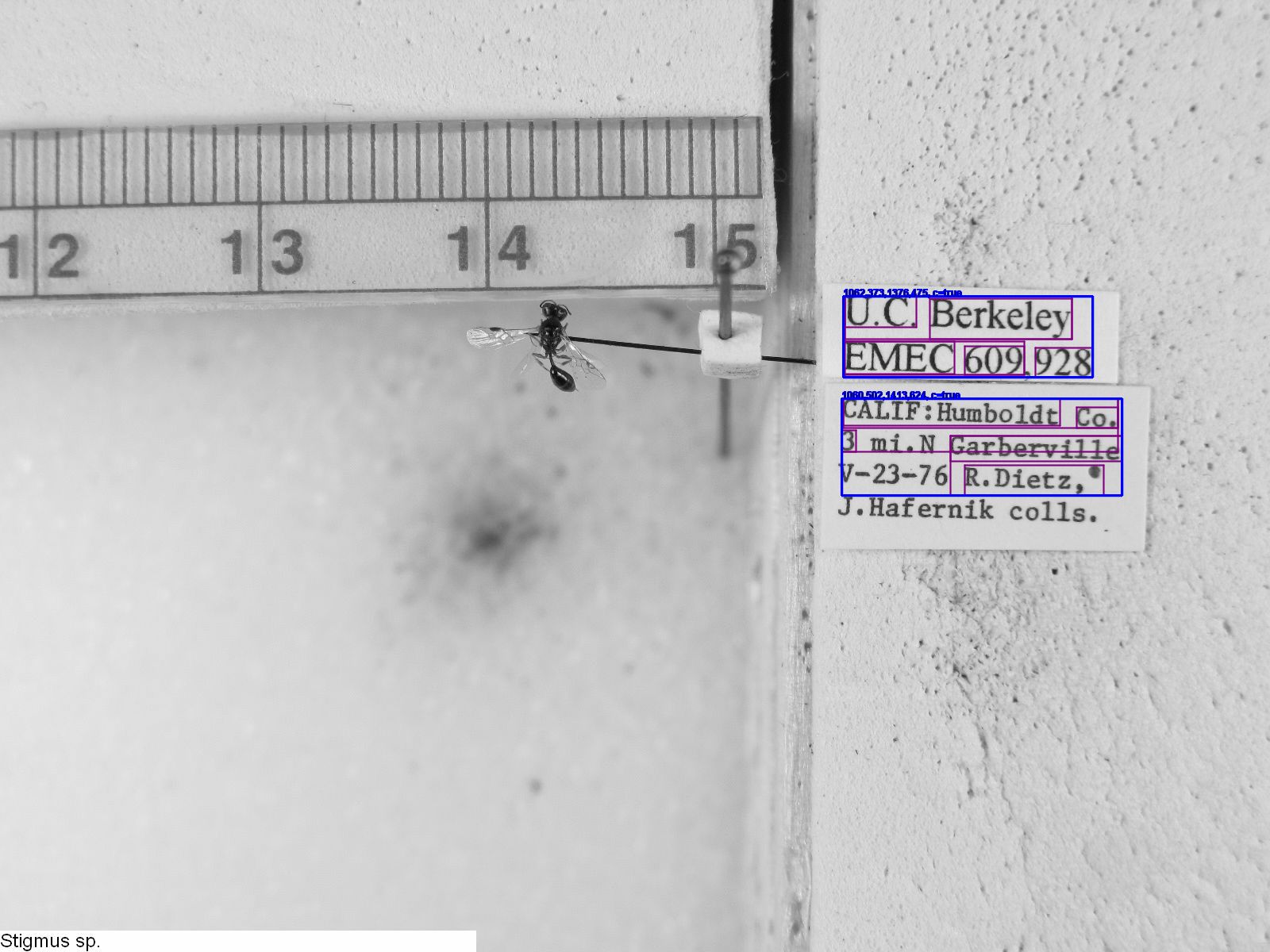
The ocrocrop method saw the greatest improvement in cases where the naive OCR pass was effective at identifying word bounding boxes, but ineffective at translating their contents into words. Taking EMEC609928_Stigmus_sp, the case of greatest improvement (naive: 18.9%, ocrocrop: 70.5%), we see that all words on the labels except for the collector name were recognized as words (in purple), making the cropped label images (in blue) good representatives of the actual labels on the image.
The cropped image was more easily processed by our OCR image, so that we may compare the naive version of the second label:
CALIF:Hunbo1dt Co. ;‘ ~ 3 m1.N' Garbervflle ,::f< '_- ' v—23~75 n.n1e:z.' 9 ._ ’
with the ocrocrop version of the second label:
CALIF:Humboldt Co. 3 mi.N Garberville V-23-76 R.Dietz,'
One of the problems with the OCR-based pre-processing which may be hidden by the scores is that many labels are entirely missed by the ocrocrop if the first, naive OCR pass failed to identify any words at all on the label. In cases such as EMEC609651_Cerceris_completa, the determination label was not cropped (indicated by blue rectangles) because no words (purple rectangles) were detected by the original. As a result, while the ocrocrop OCR is an improvement over the naive OCR (6.6% vs. 6.5%), substantial portions of text on the image are unimproved because they are unattempted.
There are two possible ways to solve this problem. One is to abandon the ocrocrop model entirely, switching back to a computer vision approach -- either by programmatically locating rectangles on the image (as Phuc Nguyen demonstrated) or by asking humans to identify regions of interest for OCR processing (as demonstrated by Jason Best in Apiary and by Paul and Robin Schroeder in ScioTR). The other option is to improve the naive OCR -- perhaps by swapping out the engine (e.g. use ABBY instead of Tesseract), perhaps by using a different image pre-processor (like ocropus's front-end to Tesseract), perhaps by re-training Tesseract.
I suspect that a computer vision approach to extracting entomology labels (or similar pieces of paper photographed against a noisy background) will provide a more effective eventual solution than the ocrocrop method. Nevertheless, the ocrocrop "bang it with a rock until it works" approach has a lot of potential to take entomology-style OCR to bad from worse.
[*]In addition to the difficulties typical of specimen labels--mix of typefaces, handwritten material, typewritten material, text inventory with few overlaps with a dictionary of literary English--the entomology dataset contained additional challenges. Difficulties included the following:
- Images containing specimens and rulers as well as labels.
- Labels casually arranged for photography, so that text orientation was not necessarily aligned.
- Labels photographed against a background of heavily pin-pricked styrofoam rather than a black or neutral background.
- 3-d images including what appear to be shadows, which soften the contrast differences around borders.