This is a transcript of my talk at the iDigBio Augmenting OCR Hackathon, presenting preliminary results of my efforts before the event.
For my preliminary work, I tried to improve the inputs to our OCR process through looking at the outputs of a naive OCR.
One of the first things that we can do to improve the quality of our inputs to OCR is to not feed them handwriting. To quote Homer Simpson, "Remember son, if you don't try, you can't fail." So let's not try feeding our OCR processes handwritten materials.
To do this, we need to try to detect the presence of handwriting. When you try to feed handwriting to OCR, you get a lot of gibberish. If we can detect handwriting, we can route some of our material to "humans in the loop" -- not wasting their time with things we could be OCRing. So how do we do this?
My approach was to use the outputs of [naive] OCR to detect the gibberish it produces when it sees handwriting to try to determine when there was handwriting present in the images. The first thing I did before I started programming, was classifying OCR output from the lichen samples by visual inspection: whether I thought there was hand writing present or not, based on looking at the OCR outputs. Step two was to automate the classifications.
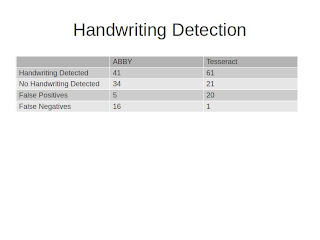
I tried this initially on the results that came out of ABBY and then the results that came out of Tesseract, and I was really surprised by how hard it was for me as a human to spot gibberish. I could spot it, but in a lot of cases -- ABBY does a great job of cleaning up its OCR output -- so in a lot of cases, particularly the labels that were all printed with the exception of some species name that was handwritten, ABBY generally misses those. Tesseract, on the other hand, does not produce outputs that are quite as clean.
So the really interesting thing about this to me is that while we were able to get 70-75% accuracy on both ABBY and Tesseract, if you look at the difference between the false positives that come out of ABBY and Tesseract and the false negatives, I think there is some real potential here for making a much more sophisticated algorithm. Maybe the goal is to pump things through ABBY for OCR, but beforehand look at Tesseract output to determine whether there is handwriting or not.
The next thing I did was try to automate this. I just used some regular expressions to look for representative gibberish, and then based on the number of matches got results that matched the visual inspection, though you do get some false positives.
The next thing I want to do with this is to come up with a way to filter the results based on doing a detection on ABBY [output] and doing a detection on Tesseract [output].
The next thing that I wanted to work on was label extraction.
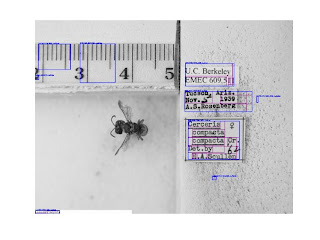
We're all familiar with the entomology labels and problems associated with them.
So if you pump that image of Cerceris through Tesseract, you end up with a lot of garbage. You end up with a lot of gibberish, a lot of blank lines, some recognizable words. That "Cerceris compacta" is, I believe, the result of a post-digitzation process: it looks like an artifact of somebody using Photoshop or ImageMagick to add labels to the image. The rest of it is the actual label contents, and it's pretty horrible. We've all stared at this; we've all seen it.
So how do you sort the labels in these images from rulers, holes in styrofoam, and bugs? I tried a couple of approaches. I first tried to traverse the image itself, looking for contrast differences between the more-or-less white labels and their backgrounds. The problem I found with that was that the highest contrast regions of the image are the difference between print and the labels behind the print. So you're looking for a fairly low-contrast difference--and there are shadows involved. Probably, if I had more math I could do this, but this was too hard.
So my second try was to use the output of OCR that produces these word bounding boxes to determine where labels might be, because labels have words on them.
If you run Tesseract or Ocropus with an "hocr" option, you get these pseudo-HTML files that have bounding boxes around the text. Here you see this text element inside a span; the span has these HTML attributes that say "this is an OCR word". Most importantly, you have the title attribute as the bounding box definition of a rectangle.
If you extract that and re-apply it to an image, you see that there are a lot of rectangles on the image, but not all the rectangles are words. You've got bees, you've got rulers; you've got a lot of random trash in the styrofoam.
So how do we sort good rectangles from bad rectangles? First I did a pass looking at the OCR text itself. If the bounding box was around text that looked like a word, I decided that this was a good rectangle. Next, I did a pass by size. A lot of the dots in the stryofoam come out looking suspiciously word-like for reasons I don't understand. So if the area of the rectangle was smaller than .015% of the image, I threw it away.
The result was [above]: you see rectangles marked with green that pass my filter and rectangles marked with red that don't. So you get rid of the bee, you get rid of part of the ruler -- more important, you get rid of a lot of the trash over here. [Pointing to small red rectangles on styrofoam.] There are some bugs in this--we end up getting rid of "Arizona" for reasons I need to look at--but it does clean the thing up pretty nicely.
Question: A very simple solution to this would be for the guys at Berkeley to take two photographs -- one of the bee and ruler, one of the labels. I'm just thinking how much simpler that would be.
Me: If the guys in Berkeley had a workflow that took the picture--even with the bee--agaist a black background, that would trivialize this problem completely!
Question: If the photos were taken against a background of wallpaper with random letters, it couldn't be much worse than this [styrofoam]. The idea is that you could make this a lot easier if you would go to the museums and say, we'll participate, we'll do your OCRing, but you must take photographs this way.
Me: You're absolutely right. You could even hand them a piece of cardboard that was a particular color and say, "Use this and we'll do it for you, don't use it and we won't." I completly agree. But this is what we're starting with, so this is what I'm working on.
The next thing is to aggregate all those word boxes into the labels [they constitute]. For each rectangle, look at all of the other rectangles in the system, expand them both a little bit, determine if they overlap, and if they do, consolidate them into a new rectangle, and repeat the process until there are no more consolidations to be done. [Thanks to Sara Brumfield for this algorithm.]
If you do that, the blue boxes are the consolidated rectangles. Here you see a rectangle around the U.C. Berkeley label, a rectangle around the collector, and a pretty glorious rectangle around the determination that does not include the border.
Having done that, you want to further filter those rectangles. Labels contain words, so you can reject any rectangles that were "primitives" -- you can get rid of the ruler rectangle, for example, because it was just a single [primitive] rectangle that was pretty large.
So you make sure that all of your rectangles were created through consolidation, then you crop the results. And you end up automatically extracting these images from that sample -- some of which are pretty good, some of which are not. We've got some extra trash here, we cropped the top of "Arizona" here. But for some of the labels -- I don't think I could do better than that determination label by hand.
Then you feed the results back into Tesseract one by one, then we combine the text files in Y-axis order to produce a single file for all those images. (Not something that's a necessary step, but that does allow us to compare the results with the "raw" OCR.) How did we do?
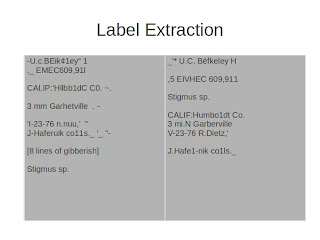
This is a resulting text file -- we've got a date that's pretty recognizable, we've got a label that's recognizable, and the determination is pretty nice.
Let's compare it to the raw result. In the cropped results, we somehow missed the "Cerceris compacta", we did a much nicer job on the date, and the determination is actually pretty nice.
Let's try it on a different specimen image.
We run the same process over this Stigmus image. We again find labels pretty well.
When we crop them out, the autocrop pulls them out into these three images.
Running those images through OCR, we get a comparison of the original, which had a whole lot of gibberish.
The original did a decent job with the specimen number, but the autocrop version does as well. In particular, for this location [field], the autocrop version is nearly perfect, whereas the original is just a mess.
My conclusion is that we can extract labels fairly effectly by first doing a naive pass of OCR and looking at the results of that, and that the results of OCR over the cropped images is less horrible than running OCR over the raw images -- though still not great.
[2013-02-15 update: See the results of this approach and my write-up of the iDigBio Augmenting OCR Hackathon itself.]